JCTC Cover: Uni-Dock, GPU-Accelerated Docking for Ultra-large Virtual Screening
Now available for free on cloud
June 15, 2023 – In a recent cover article, "Uni-Dock: GPU-Accelerated Docking Enables Ultralarge Virtual Screening," published in the Journal of Chemical Theory and Computation, DP Technology has introduced Uni-Dock, a GPU-accelerated high-performance molecular docking engine.
This technology allows an acceleration of molecular docking calculations up to 1,600 times faster than a single-core CPU on an NVIDIA V100 GPU, while preserving computational accuracy.
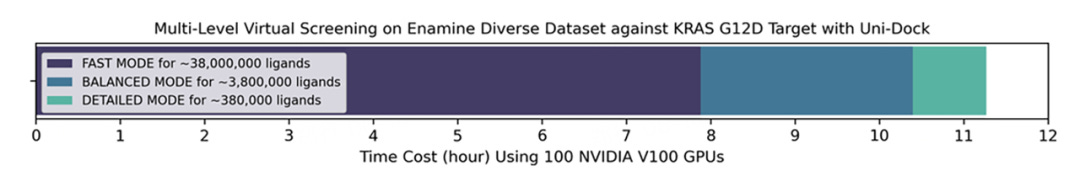
Leveraging Uni-Dock, the research team successfully completed a multistage virtual screening of 38.2 million compounds from the Enamine Diverse REAL drug database on the KRAS G12D target within just 11.3 hours, using a cluster of 100 NVIDIA V100 GPUs. The screening's average speed exceeded 37,000 molecular docking computations per GPU per hour, which substantially reduces the time and cost needed for ultra-large scale virtual screenings, thereby enabling efficient exploration of extensive chemical spaces during the early stages of new drug development.
Methodology
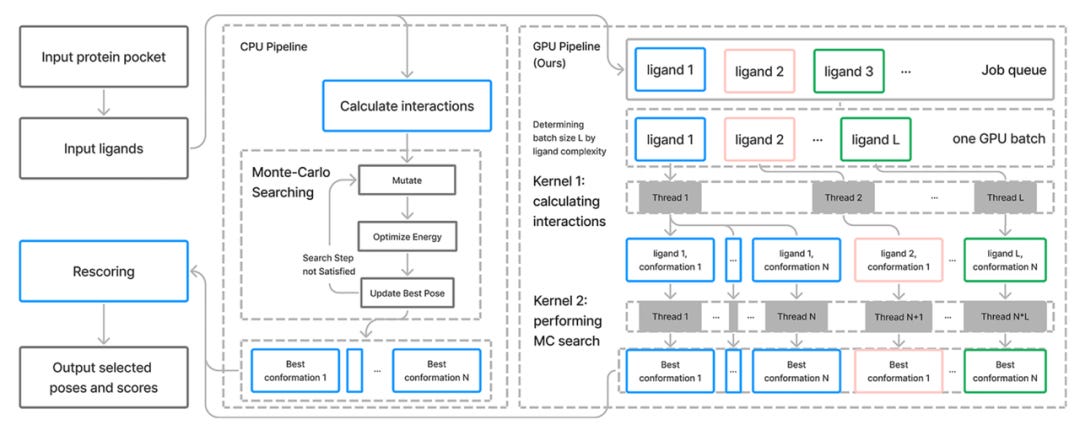
Traditional molecular docking follows a set process: A global search of protein-ligand complex conformation is conducted using the Monte Carlo (MC) method, factoring in ligand rotatable bond dihedral angles and ligand positions. The energy and force of the ligand in the current complex conformation is calculated, followed by local optimization using the BFGS algorithm to obtain the locally most stable conformation of the complex. This process is repeated until all search steps are exhausted, and the protein-ligand complex conformation with the lowest energy is returned.
Uni-Dock effectively accelerates the docking of single molecules by simultaneously launching multiple conformation search threads for a single ligand in a GPU (Kernel 2 in the graph below), reducing the computational volume per search thread by decreasing the MC iteration number for each thread. It also simultaneously launches multiple ligand docking computations to make full use of the GPU's computational capabilities (Kernel 1), dynamically assigns parallel ligand numbers based on GPU memory space, and aims to maximize ligand throughput to offset the additional cost of launching computational kernels.
Through computational logic optimization, some calculations generating massive information are transferred to the GPU, reducing host-device data transfer. Uni-Dock converts some calculations with lower precision requirements to single-precision calculations to accelerate computation and reduce GPU memory usage. Asynchronous mechanisms balance the time for CPU file reading/writing and GPU computation, and GPU memory is intelligently scheduled and dynamically allocated to fully utilize GPU computational performance across various GPU models.
Performance over Incumbant Softwares
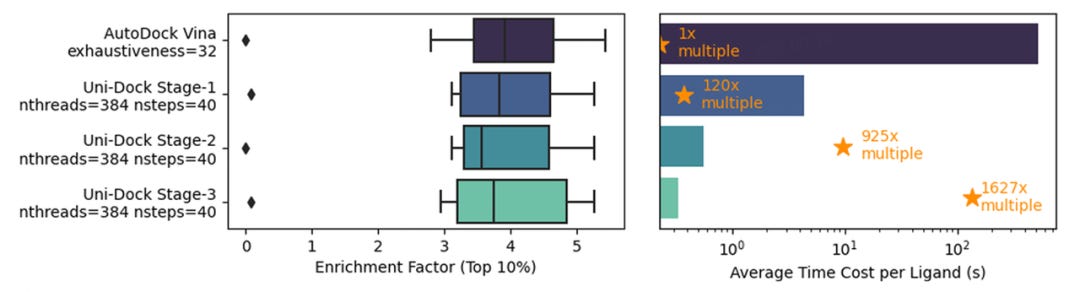
With comparable precision to AutoDock Vina (exhaustiveness=32), Uni-Dock achieves speed enhancements of 120x, 925x, and 1627x in three optimization stages. The team compared Uni-Dock with two other GPU-accelerated molecular docking software, Autodock-GPU and Vina-GPU, in terms of docking efficiency and accuracy, defining three computational complexity levels as Fast Mode, Balanced Mode, and Detailed Mode. In terms of efficiency, Uni-Dock was over 10 times faster than both competitors, with docking speeds of approximately 0.10s/ligand, 0.32s/ligand, and 0.38s/ligand across the three modes. In terms of accuracy, Uni-Dock and Vina-GPU, both based on AutoDock Vina, had comparable precision,
DP Tech's research and development team set a new performance benchmark for the latest Uni-Dock high-performance molecular docking engine, maintaining comparable accuracy across all optimization stages to the existing AutoDock Vina 1.2 engine. Using eight targets in the DUD-E database, the performance was measured based on the enrichment capability of AutoDock Vina 1.2 (with the settings: exhaustiveness=32, Vina scoring function, semi-flexible docking). The results showed that Uni-Dock achieved an impressive 1627-fold acceleration over AutoDock Vina when using an NVIDIA V100 32G GPU, compared to a single Intel® Xeon® Platinum 8269CY (Cascade Lake) 2.5 GHz CPU core.
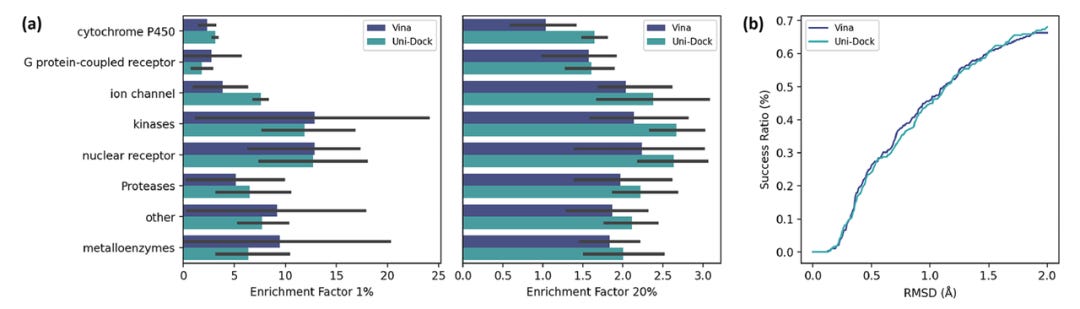
To validate Uni-Dock's multidimensional accuracy, the team compared the screening and docking power of Uni-Dock and AutoDock Vina across 102 protein targets in the DUD-E dataset (categorized into eight types) and 285 protein-ligand complexes in the CASF-2016 dataset. This comparison reaffirmed that Uni-Dock consistently matched the computational precision of AutoDock Vina at every level.
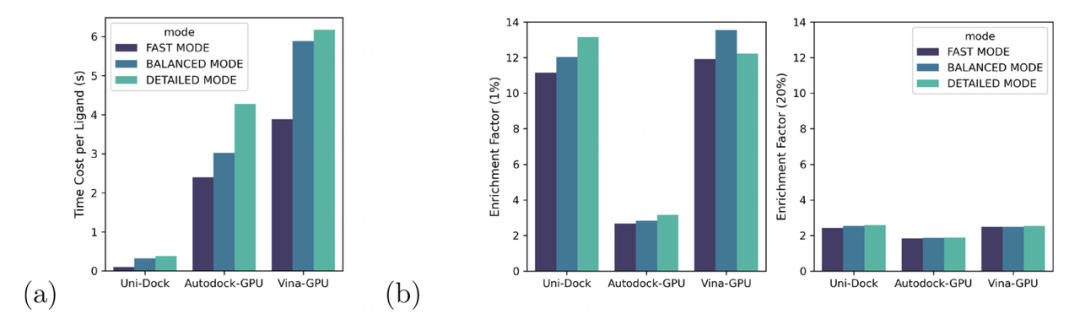
Additionally, Uni-Dock outperformed two other GPU-accelerated molecular docking software, Autodock-GPU and Vina-GPU, in both efficiency and accuracy. For an equitable comparison, three computational complexity levels were defined as Fast Mode, Balanced Mode, and Detailed Mode. In all categories, Uni-Dock exceeded its competitors by over tenfold, with docking speeds of approximately 0.10s/ligand, 0.32s/ligand, and 0.38s/ligand respectively.
Further tests demonstrated Uni-Dock's outstanding ability to linearly scale with the number of GPUs and effectively adapt to GPUs of different models and architectures. This ensures efficient deployment on varied computational resources and optimal utilization of large-scale clusters for high-throughput virtual screening.
Case Studies
DP Tech's team conducted a virtual screening on the KRAS G12D target using the Enamine Diverse REAL drug database with Uni-Dock. The database, consisting of about 38.2 million molecules, was screened using a stratified approach to balance speed and accuracy. The screening took just over 11 hours with an average rate of more than 37,000 molecular dockings per GPU hour, highlighting Uni-Dock's efficient screening capabilities.
DP Tech's Uni-Dock engine fully leverages the power of GPU parallel computing and memory space to achieve over 1600 times the acceleration ratio of AutoDock Vina, with comparable precision. It outpaces other GPU-accelerated molecular docking engines by over tenfold, achieving a molecular docking efficiency of 0.1s/ligand, and can complete virtual screening of more than 38.2 million molecular databases in less than 12 hours.
Available for FREE on Cloud-native Bohrium® Notebook and more
DP Technology has announced that Uni-Dock is now freely available to academic users! Following the usage agreement, researchers can obtain the latest release of Uni-Dock from DP Technology's GitHub repository here and use the accelerated capabilities of Uni-Dock to propel their scientific research forward.
If you're a beginner eager to learn how to use Uni-Dock, from mass submission of screening tasks to result recovery analysis, step-by-step instructions are available on the Bohrium® Notebook. Uni-Dock use-cases can be accessed here and guide users through the process.
If you'd like to conduct a virtual screening task with Uni-Dock but do not have access to a GPU machine, DP Technology also offers its Launching platform to external users. Simply submit your virtual screening job here, and the Launching platform will automatically allocate computing resources to your task, swiftly completing the virtual screening and delivering the results.
In addition, if you wish to visualize and analyze the binding poses of protein-ligand complexes, conduct further evaluation and analysis tasks such as protein structure prediction, MM-GB/PBSA, free energy perturbation (FEP) calculations, molecular property prediction, and antibody humanization design and property prediction, DP Technology offers the Hermite® drug computational design platform. Available at hermite.dp.tech, Hermite® provides a one-stop drug design solution.
Hermite® brings a novel, web-based, interactive molecular display experience for drug development scientists. It facilitates cross-window intelligent collaboration and offers diverse molecule display and operation features, making it easier for users to view, analyze, and share protein, drug molecules, and their simulation data. Hermite® supports both local and cloud-based private deployments, further enhancing its versatility and usability.
About DP Technology
DP Technology, Ltd. is an AI research and development company aspiring to solve the biggest problems in science. DP Technology is founded on the fact that our current scientific tools and computing methods have not been adequate in deepening our understanding of the microscopic world. Specifically, they have failed in predicting the behavior of complex systems at multi-scale levels. This has resulted in prolonged stagnation in the development of new materials, new drugs, new semiconductor frameworks, and beyond. We believe artificial intelligence has the potential of solving these imminent global challenges and the best way to do it is by combining the best practices of AI, scientific computing, and cloud HPC, designing innovative algorithms and software, and connecting cutting-edge technologies with real challenges in industry.